Student-facing materials
Lisa Smith and Jurriaan Ton
Learning objectives
Aims
- To understand how the expression of a chosen gene can be visualised using a reporter construct
- To determine how different environmental signals and interactions between them can affect gene expression
- To understand applications of reporter genes
At the end of this practical, you should be able to:
- Design an experiment to test a simple hypothesis
- Perform mathematical calculations for chemicals and buffers
- Use a pipette to measure volumes of liquid accurately
- Explain the importance of controls in an experiment and identify the controls in an experiment
- Use a plant reporter line to test a given hypothesis
- Describe the effects of specific signals on the expression of a gene involved in biotic response
- Write your results in a scientific format
Introduction
The plant’s innate immune system
Plants rely strongly on inducible defence mechanisms for their survival. These defences become activated upon attack by harmful organisms, such as microbial pathogens or herbivorous insects, and are part of the plant’s innate immune system. This system regulates a wide range of different chemical and physical defence barriers and is largely controlled by the hormone salicylic acid (SA; Pieterse et al., 2009). Salicylic acid is mostly activated when plants are attacked by biotrophic pathogens (Figure 1), which parasitise living plant cells. The expression of SA-inducible defence genes is commonly used as a marker for the activity of the plant immune system.
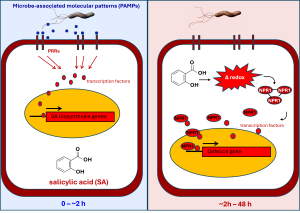
Figure 1: The signalling pathway controlling SA-inducible defence gene expression is elicited after recognition of ‘microbe-associated molecular patterns’ (MAMPs). MAMPs are small chemicals that are not produced by plants, such as breakdown products of fungal cell walls or peptides within the flagella of bacteria. Plants have evolved specific pattern recognition receptors (PRRs) that can detect these MAMPs. Within minutes after attack, PRR-mediated MAMP perception activates downstream transcription factors that in turn activate nuclear genes encoding for SA biosynthesis enzymes. As a consequence, more SA is produced, and cellular concentrations of the hormone start to rise. Via mechanisms that are still poorly understood, the increasing SA levels induce rapid fluctuations in the cytoplasmic redox state (i.e. the concentration of reactive electrons), which partially reduce complexes of the defence regulatory protein NPR1 into monomers that can move to the nucleus. Once NPR1 enters the nucleus, the protein starts to recruit transcription factors that initiate defence gene transcription.
The signalling pathway controlling SA-inducible defence gene expression is complex and involves initial recognition of ‘microbe-associated molecular patterns’ (MAMPs) by pattern recognition receptors (PRRs). MAMPs are small chemicals that are not produced by plants, such as breakdown products of fungal cell walls, which plants use to detect that they are under attack by a microbial pathogen. Perception of MAMPs triggers a rapid cellular signalling cascade within minutes to hours after attack, which activates transcription factors that migrate to the nucleus, where they bind to gene promoters of genes involved in biosynthesis of SA. As a consequence, SA accumulates in the cytoplasm.
Salicylic acid versus Aspirin
SA acts as an analgesic and immuno-suppressant in animals. It is the active ingredient in willow bark, which used to be chewed as a pain remedy. The chemical structure of SA is very similar to that of aspirin, which is acetylsalicylic acid (Figure 2). Paradoxically, while SA functions as an immune stimulant in plants, SA and aspirin act as immuno-suppressants in animals. Whether aspirin has the same stimulatory effects as SA in plants, or whether it can act as an immuno-suppressant, can be investigated during this practical. The pPR1:GUS Arabidopsis reporter line can be used as a means to visualize activity of the SA-inducible immune system after spraying the leaves with a solution containing SA and/or aspirin (available in the teaching lab).
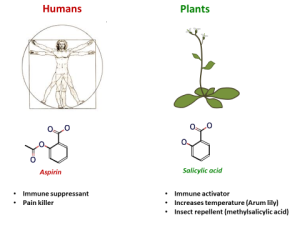
Figure 2: Chemical structures and roles of salicylic acid (left) and aspirin (right) on animals and plants, respectively
Plectosphaerella cucumerina: a soil fungus gone rogue
The ascomycete fungus Plectosphaerella cucumerina is commonly found in soil. However, some strains of this fungus have evolved the ability to infect plant tissues. Figure 3 shows a time course of disease development of this fungus at different time-points after application of a droplet containing P. cucumerina spores. The SA response is particularly effective against biotrophic pathogens that parasitize living cells. Necrotrophic pathogens, which kill plant cells and use the dead tissue as substrate to colonize the plant, are insensitive to SA-dependent defences. Most pathogens start their infection cycle with a biotrophic lifestyle and switch after a couple of days to a necrotrophic lifestyle. They do this to escape the SA-dependent immune response that is activated during the earlier (biotrophic) stage of their infection cycle. Whether P. cucumerina activates SA-dependent defences during its infection cycle is something that you can test during this practical. You can use the pPR1:GUS Arabidopsis reporter line to test whether the SA response is activated at the sites of fungal spore application. A spore suspension of P. cucumerina is available in the teaching lab, and you can use pipettes to apply 5µL droplets to selected leaves.

Figure 3: Time course of symptom development by Plectosphaerella cucumerina. Fully expanded leaves from Arabidopsis thaliana (accession Col-0) were infected by applying 5 µL droplets containing 5 x 106 spores/mL . Photographs were taken at 5-day intervals.
Does abiotic stress weaken the SA-controlled immune system?
The efficiency of the SA-controlled plant immune system can be influenced by abiotic environmental conditions, such as temperature, water and nutrient availability, and light intensity (Pieterse et al., 2009). For instance, the plant hormone abscisic acid (ABA) is a central regulator of plant responses to changes in abiotic growth conditions, such as drought and salt stress, but can have cross-effects on plant resistance to diseases (Ton et al., 2009). It is therefore likely that drought- or salt-exposed plants respond differently to pathogen attack, which can have far-reaching consequences for maintaining crop yields in a warming climate. For example, soil salinity is increasing in many parts of the world due to increased irrigation. Whether salt stress can suppress the SA-controlled immune system in Arabidopsis can be tested during this practical. For instance, you can apply salt solution to roots of the pPR1:GUS Arabidopsis reporter line and test whether the SA response is suppressed upon spraying the leaves with a SA solution. Both salt and SA solutions will be available in the teaching lab.
Research Questions
Each group should choose ONE of the following questions to test:
- Do aspirin and salicylic acid (SA) have similar effects on PR1 gene expression?
- Does the fungus Plectosphaerella cucumerina induce SA signalling and PR1 gene expression?
- Does salt stress affect SA-induced PR1 gene expression?
Background information for key materials
Plant material
Two transgenic GUS reporter lines of Arabidopsis thaliana are available. The first line carries a transgene in which the promoter region of a SA-inducible (pPR1::GUS; stock number N6357 from the Nottingham Arabidopsis Stock Centre) gene is fused to the beta-glucuronidase (GUS) reporter gene from E. coli. If these plants accumulate SA, the cells will activate the GUS reporter gene, which causes accumulation of stable GUS enzyme. The extent of GUS activity can then be measured by incubating leaves in staining solution containing 5-bromo-4-chloro-3-indolyl beta-D-glucuronide (X-GlcA), which is converted into a blue precipitant in the presence of GUS enzyme. The “blueness” of the tissue is directly proportional to the level of SA-induced gene expression. The second transgenic GUS reporter line is a constitutive expressor of GUS (p35S::GUS). You will also have some wildtype Col-0 plants (stock number N1092 from the Nottingham Arabidopsis Stock Centre) without a GUS reporter. All plants have been grown in advance for three weeks in a growth chamber at 22ºC with 10 hours of light per day.
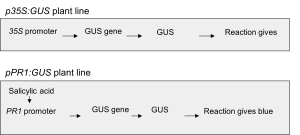
Figure 4: Flow diagram of signalling components in the two GUS reporter lines used in this practical.
Infection assays with Plectosphaerella cucumerina.
Plectosphaerella cucumerina is an ascomycete fungus that can survive saprophytically in soils and on artificial growth media. The pathogenicity assays will be carried out as described by Ton and Mauch-Mani (2004). Plectosphaerella cucumerina will be grown from frozen glycerol stocks on agar plates containing 19.5 g/L potato dextrose agar (Difco, Detroit) at room temperature for at least 10 days. Spores can be collected by adding 5 – 10 mL 10 mM MgSO4 to the agar plate and gently rubbing the agar surface with a spatula. Spore density of the collected suspension will be adjusted to 5 x 106 spores/mL. 10 mM MgSO4 is used as an osmotic buffer. Inoculation is typically performed by applying 3 µL droplets of the inoculum onto fully expanded leaves. After inoculation, plants are kept at high humidity to stimulate disease. Disease symptoms will appear from 4 to 6 days after treatment.
Schedule of work
Day 1
This first day of the practical involves two introductory lectures. Since not all of you have done the Molecular & Cell Biology course in the 1st Semester, the first lecture will re-iterate the basic principles of eukaryotic gene regulation, which are necessary to understand the working of the reporter genes that our practical is based on. The second lecture will be general introduction to the practical, which will be based around experiments with transgenic plants carrying a reporter construct of a plant defence gene. At the end of the second lecture, you will need to choose one of the three available research question that you can investigate in the practical.
Day 2
1) Write out the research question in the form of the hypothesis you will be testing
2) Regardless of research question, you have the following plant genotypes to test your hypothesis:
- 2 pots of 3-4 wildtype plants (Col-0)
- 2 pots of 3-4 plants that constitutively express GUS (p35S::GUS)
- 4 pots of 3-4 plants with an inducible pPR1::GUS reporter
3) You also have the following treatment solutions available to test your hypothesis:
-
- distilled water
- 5 mM aspirin in water
- 5 mM SA in water
- 5 mM SA in 10 mM MgSO4
- 300 mM NaCl
- Plectosphaerella cucumerina inoculum in 10 mM MgSO4
- 10 mM MgSO4
4) Decide which treatment(s) you will apply to each of your plants to test your hypothesis. You will get 4 pots with the pPR1::GUS reporter lines, 2 pots of the wild-type (Col-0) and 2 pots of the p35S::GUS lines. Once you have designed the experiment, indicate the treatment(s) you will give each pot of plants in the table below. Also consider what is being tested through this combination of genotype and treatment, and which other genotype-treatment combinations you will need to compare your results to.
| Pot of Plants | Treatment | Testing what? | Compare to which other plants? |
| Wildtype 1 | |||
| Wildtype 2 | |||
| pPR1::GUS 1 | |||
| pPR1::GUS 2 | |||
| pPR1::GUS 3 | |||
| pPR1::GUS 4 | |||
| p35S::GUS 1 | |||
| p35S::GUS 2 |
6) Which plants are your controls and why?
7) If there were further plants available, what genotypes and treatments would you add to your experimental design?
8) Discuss your experimental design with a demonstrator.
9) Pipetting practice.
During this practical, only some students will need to use a pipette. The ability to use a pipette is a core skill in biology these days, therefore we want to make sure you all have the opportunity to learn how to use a pipette. A balance, beaker to pipette into, and water are provided at the end of each bay. Take your pipettes and pipette tips to the balance, and pipette the following amounts of water into the beaker, re-zeroing the balance between each volume. Record the weight of water pipetted for each student in your group and each volume in the table on the next page. If no other groups are waiting for the balance, you can try pipetting other volumes too. If you are unsure at any step, make sure you ask a demonstrator for help.
What was the weight of water pipetted by each student for each volume?
| Volume | 1000 µl | 450 µl | 200 µl | 78 µl | 20 µl | 12.5 µl |
| Pipette | P1000 | P1000 | P200 | P200 | P20 | P20 |
| Student 1 | ||||||
| Student 2 | ||||||
| Student 3 | ||||||
| Student 4 |
10) Which student in your group was most accurate with their pipetting? Are the pipettes accurate to within 1% of the expected volumes?
11) You are now ready to treat your plants. Collect your plants from the labelled trays at the front of the room. Make sure you also take one plastic label per plant. Label the plants IMMEDIATELY with your group names and the plant genotype (wildtype or Col-0, pPR1::GUS or p35S::GUS) so that you don’t mix up your plants. They all look the same so if you don’t label them, you won’t know which is which! It might be a good idea if individual group members collect plants from only one genotype.
Each treatment solution should be applied in the following ways:
- distilled water – pipetted droplets, root drench or sprayed
- 5 mM aspirin in water – sprayed
- 5 mM SA in water – sprayed
- 5 mM SA in 10 mM MgSO4 – pipetted droplets
- 300 mM NaCl – root drench
- Plectosphaerella cucumerina inoculum – pipetted droplets
- 10 mM MgSO4 – pipetted droplets
You will find the treatment solutions in trays at the front of the bays, with the exception of the NaCl and water root drenches which are already in the trays for putting the plants in at the front of the bay.
For spray treatments: take a spray bottle from the front of the bay, label your pot of plants with the treatment you are going to apply, treat your plants by spraying them so that the leaves are at least covered in small droplets, then return the spray bottle.
For root drenches: put your labelled pot of plants in the respective tray.
For pipetted drops: use a P20 pipette set to 3 µl to apply droplets of the solution directly onto your leaves. It is up to you to decide how many leaves to treat and how many droplets to apply to each leaf. You may wish to mark which leaves of the plants you treated by using a thick black lab marker if only treating a subset of the leaves with pipetted droplets. If you are unsure of how to set your pipette, ask a demonstrator for help.
12) Go ahead and treat your plants as you planned. Make sure all your pots are fully labelled with your group names, the genotype and treatment (not just a number), and readily identifiable before you put them into the appropriate trays at the front of the lab. The plants will be returned to the growth room for the next week.
13) Remember to wash your hands on the way out of the lab.
Day 3
1) Your plants are located at the front of the room. Collect all of your pots.
2) Examine your plants to see if any of the treatments has given a visible phenotype. Fill in the table below:
| Plant | Treatment | Phenotypic observations |
| Wildtype 1 | ||
| Wildtype 2 | ||
| pPR1:GUS 1 | ||
| pPR1:GUS 2 | ||
| pPR1:GUS 3 | ||
| pPR1:GUS 4 | ||
| p35S:GUS 1 | ||
| p35S:GUS 2 |
3) If you do note phenotypic differences, why do you think they may have arisen? Discuss with a demonstrator and fill in your answer below.
4) Once you have finished examining your plants, you can move on to the GUS staining. You have been provided with eight 1.5 mL aliquots per group of the GUS staining solution. We have prepared this in advance as follows.
For 20 mL of staining solution the following stock solutions were combined:
| Stock | Stock concentration | Volume |
| X-GlcA | 20 mg/mL in DMSO | 0.55 mL |
| NaH2PO4/Na2HPO4 | 0.5 M (pH = 7) | 4 mL |
| K4Fe(CN)6 | 25 mM | 0.2 mL |
| K3Fe(CN)6 | 25 mM | 0.2 mL |
| Triton X-100 | 10% v/v | 0.2 mL |
| dH2O (distilled water) | – | 14.85 mL |
This staining solution was prepared in advance for two reasons. Firstly, to save time during the practical, and secondly, because DMSO, K3Fe(CN)6 and K4Fe(CN)6 are toxic if ingested. As a precaution, you MUST wear gloves when handling the staining solution. If you do get staining solution on yourself, don’t panic as it will not be absorbed through your skin. However, do wash it off your skin immediately with copious soapy water to ensure you do not accidentally ingest the solution.
Even though we have prepared all the solutions in advance for you, it is important that you understand how to make up chemical solutions. Did you notice that the stock concentration of Triton X-100 is given as ‘10% v/v’ rather than as a molarity? This means that the Triton X-100 stock is a 10% solution, volume to volume, with the second volume referring to distilled water unless otherwise specified. So a stock of Triton X-100 would be made by measuring out 1 mL of this detergent (which is VERY viscous!) then adding 9 mL of dH2O and inverting gently until the Triton X-100 and water are completely mixed. Sometimes you will see the notation ‘w/v’ instead of ‘v/v’, which means that a solution is then prepared ‘weight to volume’. A weight of 1 g per mL would be a 100% w/v solution.
5) What would be another way of expressing the composition of the X-GlcA stock solution, other than ‘20 mg/mL in DMSO’?
When calculating how much of a chemical to use in preparing a solution, remember that you need to take a number of things into consideration:
- The (formula) molecular weight of the chemical, which is usually listed as MW or FW on the bottle. Remember that the units for MW are g mol-1.
- The volume you are preparing in litres (L).
- The concentration of the solution you are preparing in molarity or moles per litre (mol L-1).
The formula you need to use is n = cv where n is number of moles, c is concentration in molarity (= moles per litre), v is volume in litres. Also, n = weight / MW.
Going back to the GUS staining solution, the sodium phosphate buffer stock is prepared at pH 7.0 and 0.5 M. To prepare this buffer, two forms of sodium phosphate are combined; the monobasic form (NaH2PO4) and the dibasic form (Na2HPO4). This can be done by making 1 M solutions of each and combining the solutions in the correct ratio according to the table below to achieve the desired pH (table is for 1 L of 1 M buffer). Of course, it is always good practice to then check the pH of your solution with a pH meter!
| Final pH | Volume 1 M NaH2PO4 (mL) | Volume 1 M Na2HPO4 (mL) |
| 6.0 | 877 | 123 |
| 6.5 | 685 | 315 |
| 7.0 | 390 | 610 |
| 7.5 | 160 | 840 |
| 8.0 | 53 | 947 |
6) Given that you would use 4 mL of 0.5 M sodium phosphate buffer at pH 7.0 to make the GUS staining solution, calculate how much of each salt is needed to make the staining solution. Give your answer in g or mg and show your working.
For your calculations: NaH2PO4·2H2O, MW 155.99
Na2HPO4·7H2O, MW 268.07
7) You use 0.2 mL of 25 mM K4Fe(CN)6 to make up 15 mL of GUS staining solution. What is the final concentration of K4Fe(CN)6?
8) Consider why each component is included in the GUS staining solution. For example, why is the detergent Triton X-100 included?
9) Put gloves on before handling the tubes containing staining solution or, as is good laboratory practice, anything in the lab – you never know where someone else might have spread a toxic chemical.
10) Place collected leaves from the different lines and treatments into the staining solution using the forceps provided. Remember that there may be biological and treatment variability between leaves, so you may want to collect multiple leaves for each genotype and treatment. Ensure that your collected samples are completely submerged in the staining solution. Make sure you label each tube with your group name, the date and what your sample is (genotype and treatment).
11) Bring your samples to the front of the bay and ask the demonstrator to vacuum infiltrate the tissues in the desiccator located there. The tutor will vacuum infiltrate your tissues for approximately 5 minutes with multiple rounds of applying and releasing the vacuum.
12) Why do the leaves produce bubbles under a vacuum?
13) Check that the tissue has sunk to the bottom of each eppendorf. If not, ask the demonstrator to repeat the vacuum infiltration. Bring your samples to the front of the bay and place them in the collection tray. Your samples will be incubated at 37 °C until [Day 4].
14) Dispose of your waste plant tissue into the autoclave bags at the front of room (the clear bags with blue writing on them). Labels from the plant pots should be put in the beaker beside the autoclave bags.
15) Make sure you record how your tubes are labelled.
16) Remember to wash your hands on the way out of the lab.
Day 4
This will be a very short day. You can use the extra time to ask your demonstrator about aspects that have remained unclear and/or you can start working on your report.
1) Put gloves on (and your lab coat!). Collect your tubes from the racks at the front of the bay into an empty rack.
2) Pipette the GUS staining solution out of the 2 mL eppendorf tubes and dispose of it in the provided liquid waste containers. Dispose of the pipette tip in the provided solid waste container.
3) Using a P1000 pipette, and pipetting twice, add 1.8 mL of 70% ethanol to each eppendorf tube. Please do not overwind the pipettes – remember that a P1000 can pipette 1000 µl max. You therefore need to pipette 900 μL (0.9 mL) twice. Also, be careful not to get 70% ethanol on the outside of the tubes or your labels will come off! (Lab marker inks are generally water-resistant but not alcohol-resistant). Dispose of the pipette tip in the solid waste container provided.
4) Return your tubes to the collection tray at the front of the bay for storage until next week.
5) Remember to wash your hands before leaving the lab.
Day 5
- Examine your stained leaves to see the extent of GUS staining as indicated by blue colour. You can remove your samples from the tubes using forceps and put them between two sheets of transparent plastic to take a closer look, but remember to wear gloves as there will be some carry over of the GUS staining solution. You can label your samples by writing on the upper plastic sheet. It may also be useful to get a piece of blank paper from the front of the bay to put under your plastic sheets so you can see your leaves better. Fill in the table below with an estimate of the extent of GUS staining on a scale from 1 – 5 for each sample, with 1 being no GUS staining and 5 a completely blue leaf. You can also draw a picture of your results if you wish:
| Plant | Treatment | Extent of GUS staining (1 – 5) | Further observations |
| Wildtype 1 | |||
| Wildtype 2 | |||
| pPR1:GUS 1 | |||
| pPR1:GUS 2 | |||
| pPR1:GUS 3 | |||
| pPR1:GUS 4 | |||
| p35S:GUS 1 | |||
| p35S:GUS 2 |
2) Once you have finished examining your samples, you should dispose of them by decanting the liquid into the small beaker in your tray, putting the rest of the tissue and 1.5 mL tubes into the waste bags at the front of the bays.
3) Based on your results, what conclusions can you reach? It might be helpful to refer back to point 5 from the first week.
4) Did your controls give the expected results? If not, then why might these results have been different to expectations?
5) Did your results prove or disprove your hypothesis?
6) If the experiments were not conclusive, then what further experiments could be conducted to prove or disprove the hypothesis?
7) If you were to repeat your experiments, what would you change and why? Would you change how you quantified the GUS staining?
8) This last lab session will be followed by a summary session presenting typical results from all three experiments. For now, it is a good idea to discuss the experiments and results with other groups studying the same research question to see if you all reached the same conclusion. We also encourage you to understand the experimental design and results from all three experimental questions, which allows you to make reference to the other two research questions in your report. So, you may also wish to talk to groups who investigated a different research question and fill in the tables and boxes below as a record of the results.
Research Question 1: Do aspirin and salicylic acid (SA) have similar effects on PR1 gene expression?
| Plant | Treatment | Phenotypic observations | Extent of GUS staining (1 – 5) | Further observations |
| Wildtype 1 | ||||
| Wildtype 2 | ||||
| Wildtype 3 | ||||
| pPR1:GUS 1 | ||||
| pPR1:GUS 2 | ||||
| pPR1:GUS 3 | ||||
| p35S:GUS 1 | ||||
| p35S:GUS 2 | ||||
| p35S:GUS 3 |
Which plants were the controls and what did they each test?
Further notes:
Research Question 2: Does the fungus Plectosphaerella cucumerina induce SA signalling and PR1 gene expression?
| Plant | Treatment | Phenotypic observations | Extent of GUS staining (1 – 5) | Further observations |
| Wildtype 1 | ||||
| Wildtype 2 | ||||
| Wildtype 3 | ||||
| pPR1:GUS 1 | ||||
| pPR1:GUS 2 | ||||
| pPR1:GUS 3 | ||||
| p35S:GUS 1 | ||||
| p35S:GUS 2 | ||||
| p35S:GUS 3 |
Which plants were the controls and what did they each test?
Further notes:
Research Question 3: Does salt stress affect SA-induced PR1 gene expression?
| Plant | Treatment | Phenotypic observations | Extent of GUS staining (1 – 5) | Further observations |
| Wildtype 1 | ||||
| Wildtype 2 | ||||
| Wildtype 3 | ||||
| pPR1:GUS 1 | ||||
| pPR1:GUS 2 | ||||
| pPR1:GUS 3 | ||||
| pPR1:GUS 4 | ||||
| p35S:GUS 1 | ||||
| p35S:GUS 2 | ||||
| p35S:GUS 3 |
Which plants were the controls and what did they each test?
Further notes:
Make sure you understand the design and results from at least your experiment before you leave the laboratory. If you are unclear on any points, please discuss them with the demonstrators. Also, please remember to wash your hands before leaving the lab.
Day 6
This final day of the practical is a discussion and Q&A session, where we will re-iterate the results of each research question and answer remaining questions you may have about your own results and your report.
References/Further Reading
Pieterse CMJ, Leon-Reyes A, Van Der Ent S, Van Wees SCM, 2009. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology 5, 308-16.
Ton J, Mauch-Mani B, 2004. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J 38, 119-30.
Ton J, Flors V, Mauch-Mani B, 2009. The multifaceted role of ABA in disease resistance. Trends Plant Sci 14, 310-7.

