Resources for teachers
Emma Jones; Michelle Durrant; and Sarah Noble-Longster
This paper was key to the design of my practical. I also obtained the 13 cat mtDNA sequences of known haplotype from the first author:
Barbara Ottolini, Gurdeep Matharu Lall, Federico Sacchini, Mark A. Jobling, Jon H. Wetton, Application of a mitochondrial DNA control region frequency database for UK domestic cats, Forensic Science International: Genetics, Volume 27, 2017, Pages 149-155, ISSN 1872-4973. https://doi.org/10.1016/j.fsigen.2016.12.008.
The method for PCR amplification has been taken from the following paper:
RA Grahn et al, Feline non-repetitive mitochondrial DNA control region database for forensic evidence, Forensic Science International: Genetics, Volume 25, 2010, Pages 33-42. doi: 10.1016/j.fsigen.2010.01.013
https://pmc.ncbi.nlm.nih.gov/articles/PMC2921450/
Results from producing restriction maps of the four sequences:
-
- AlfI – doesn’t cut any; not useful
- ApoI – cuts all four at 469; not useful
- BccI – cuts all at 243; not useful
- BsaA1 – cuts #2, 3 and 4 at 418; would single out suspect 1
-
- BseYI – cuts all at 386; not useful
- BsmA1 – cuts ~1 and 2 at 414; cuts #3 at 45; doesn’t cut #4; would single out suspect 4, and possibly suspect 3
- ClaI – doesn’t cut any; not useful
- DraII – cuts #1 and 2 once; cuts 3 and 4 twice; distinguishes 1 or 2 from 3 or 4
- HaeII – doesn’t cut any; not useful
- HindIII – doesn’t cut any; not useful
- PmeI – doesn’t cut any; not useful
- SmlI – cuts all at 85; not useful
- XhoI – cuts all at 85; not useful
Students should therefore set up three separate reactions with BsaA1, BsmA1 and DraII (alternative enzyme that cuts at the same site as DraII is EcoO109I). The results will be a mess if students perform all digests in one tube.
Band sizes expected from each restriction enzyme for each suspect cat:
| BsmA1 | BsaA1 | DraII | |
| Suspect 1 | 413, 77 | 490 | 358, 132 |
| Suspect 2 | 413, 77 | 417, 73 | 358, 132 |
| Suspect 3 | 446, 44 | 417, 73 | 358, 48, 84 |
| Suspect 4 | 490 | 417, 73 | 358, 48, 84 |
Expected gel results from cutting the extracted and amplified mtDNA:
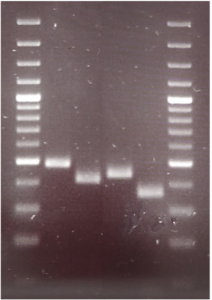
1 & 6: 100 bp-Plus GeneRuler
2: uncut fragment
3: BsaAI
4: BsmAI
5:EcoO109I

