Technical information
Janet Cronshaw; Elizabeth Alvey; Catherine Heath; Qaiser Sheikh; Melanie Stapleton; and Ilona Willson
Day 1
| ITEM | AMOUNT | NOTES |
| PER PAIR | ||
| Linearised pBH763 | 40 μl
(containing 2 μg plasmid digested with NotI) |
On ice
See below for details |
| 3 μg pBH750 | 15 μl at 0.2 μg/μl | On ice
See below for details |
| K699 yeast
(grown overnight in YAPD + amp, at 30°C/200 rpm until OD600nm is- 0.4-2.0) |
10 ml | On ice
See below for details in yellow universals that can be centrifuged |
| 0.1 M lithium acetate | 5 ml | |
| 1.0 M lithium acetate | 200 μl | |
| 50 % (w/v) PEG | 1 ml | |
| salmon sperm DNA (10 mg/ml)
(0.25 μm-filtered) |
100 μl | On ice |
| YAPD | 5 ml | See below for details |
| Sterile distilled water | Put a fresh bottle in drawers | |
| Sterile plastic universals | 3 | |
| Elastic band | 1 | |
| -leu plates + adenine (80 mg/L) + amp | 3 | See below for details |
| Alcohol bath & spreader | 1 | Use IMS, not ethanol |
| Vector handout | 1 per person | |
| PER BAY | ||
| Virkon pots | 1 between 2 stations | |
| EQUIPMENT | ||
| Hot blocks at 30°C and 42°C | ||
| Shaking water baths at 30°C | ||
| OTHER | ||
| Bay trays for students’ plates. Incubate at 30°C for 2-4 days then transfer to cold room |
YAPD = YPD + adenine (80 mg/L). Adenine can be added to the media before autoclaving
-leu + adenine + amp plates (0.5 L):
- Yeast synthetic drop-out medium
- Supplement w/o leucine (BIO 101 CSM-leu) 0.8 g
- Yeast nitrogen base w/o amino acids 3.33 g
- Glucose 10 g
- Adenine 40 mg
- Difco agar 10 g
Autoclave, cool and add amp before pouring.
Expected transformation results
| Transformation | 1 μg plasmid (pBH750) | Repair template | Expected leu+? | Expected NrsR? | Expected Met–? |
| 1 | Yes | Yes, linearised | Yes | Yes | Yes |
| 2 | Yes | No | Yes | No | Yes |
| 3 | H2O only (-ve control) | No | No | No | |
Plasmids
| plasmid | contains | function | result |
| pBH750
(ampR) |
CRISPR-Cas9 | Cleave DNA |
|
| Guide RNA (gRNA) for MET15 locus | Methionine synthesis (MET15) | Guides CRISPR-Cas9 to the MET15 locus and causes DNA cleavage at this site | |
| LEU2 | Leucine synthesis | Allows selection of transformants | |
| pBH763
(ampR) |
500 bp upstr + downstr met15 HR region | Homologous repair template for met15 gene | Directs repair template and therefore natMX6 to met15 gene where the DNA cleavage occurred |
| natMX6 gene
in the middle of the above met15 HR region to give met15-NatMX6-met15 cassette |
Nourseothricin (NatMx) resistance | Nourseothricin (NatMx) resistance | |
| NotI sites either side of met15-NatMX6-met15 cassette | To release a linearised repair template | Cells can be transformed with much higher efficiency using linearised (rather than circular) DNA |
Day 2
| ITEM | AMOUNT | NOTES |
| PER PAIR | ||
| YAPD + amp plates | 4 | See below for details |
| YAPD
+ nourseothricin (50 μg/ml) + amp plates |
2 | See below for details |
| -met + adenine (80 mg/L) + amp plates | 2 | See below for details |
| Numbered grids | 2 | They use these just for the grid pattern rather than the numbers |
| sgRNA design activity handout | one per person | |
| PER BAY | ||
| Control plates of K699 and K699 NatMx+ | one plate of each between two pairs | |
| EQUIPMENT | ||
| OTHER | ||
| Students’ CRISPR plates from the previous session | ||
| Rescue transformation plates on spares trolley | Grown on fake -leu + adenine + amp plates (see below) | |
| Bay boxes for YAPD and -met plates; incubate at 30°C overnight and then store at 4°C until the next session | ||
| No need for sterile Eppendorfs – they use too many and there’s amp in the plates anyway |
YAPD plates = YPD agar + adenine (80 mg/L). Adenine can be added to media before autoclaving
Autoclave, cool to 50°C and add nourseothricin (50 μg /ml) and/or amp before pouring
-met + adenine + amp plates (0.5 L):
- Yeast synthetic drop-out medium supplement w/o methionine (BIO 101 CSM-leu): 0.8 g
- Yeast nitrogen base w/o amino acids: 3.33 g
- Glucose: 10 g
- Adenine: 40 mg
- Difco agar: 10 g
Autoclave, cool and add amp before pouring.
Fake -leu + adenine + amp plates (for transformation rescue plates)
(i.e. as per recipe for -leu plates in CRISPR #1 but with drop-out without methionine and without leucine, so not actually selective) (0.5 L):
- Yeast synthetic drop-out medium supplement w/o methionine: 0.4 g
- Yeast synthetic drop-out medium supplement w/o leucine: 0.4 g
- Yeast nitrogen base w/o amino acids: 3.33 g
- Glucose: 10 g
- Adenine: 40 mg
- Difco agar: 10 g
Notes on checking growth of S. cerevisiae K699
Need for adenine
K699 is apparently an auxotroph for adenine. Check how presence of 40 mg/L adenine affects growth on complex media (YPD). NB adenine-HCl added before autoclaving, as I had trouble making up a stock solution of adenine-HCl (would not dissolve in water, even at half the concentration of Sigma’s stated solubility).
- Grew K699 O/N at 30°C/shaking in 5 ml YPD or YAPD, from a glycerol stock or a plate inoculation (both cultures grew).
- Then streaked all four cultures onto YPD + amp plates or YAPD + amp plates
- Incubated at 30°C for 2d
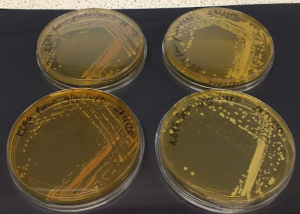
Appears that K699 can grow on YPD without adenine, but that the addition of adenine (RH plates) results in bigger colonies. So continue to add approx. 40 mg/L adenine-HCl to YPD media for growth of this strain.
Update: after a few days storage in fridge, cultures grown on YPD plates were distinctly red
OD for transformations
Protocol states that K699 should be at OD600 nm ~0.4-2.0. Test OD of overnight culture vs A16 (which also has to be in log phase for mol gen transformations in autumn).
A16 is grown as follows in autumn: 200 μl inoculum into 350 ml YPD in 2 L flask, grown at 30°C overnight at 200 rpm.
Scale down to 44 ml in 250 ml flask with 25 μl inoculum, grown at 30°C overnight at 200 rpm. Grow A16 in YPD and K699 in YAPD
OD600nm after 16 h:
A16 = 1.17
K699 = 1.06
So K699 grows to a similar OD600nm as A16, suggesting it should be OK (assuming Hansenula and Saccharomyces behave similarly in transformations). Make a note to take the flask out on the morning of the class and put on ice to avoid any further growth.
Nourseothricin concentration
Protocols state to use 50-100 μg/ml nourseothricin. Check growth of K699 (bottom row) vs a control NatMx+ strain (top row) on these concentrations:
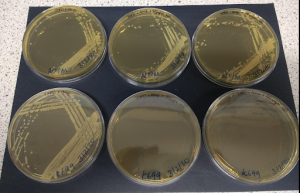
0 50 100 ug/ml
50 μg/ml is sufficient, 100 μg/ml is not detrimental to growth
Access to plasmids
Please contact Mel Stapleton (m.stapleton@sheffield.ac.uk) to make arrangements.

